How SMC proteins pack the genetic material in the cell nucleus
The entire genetic information of living organisms is stored in the form of DNA in the tiny nuclei of their cells. How a protein called Smc5/6 controls the safe packaging of genetic material in chromosomes by extruding loops from the DNA has now been revealed by an international team of researchers from Eugene Kim’s group at the Max Planck Institute of Biophysics in Frankfurt am Main (Germany) and from the Karolinska Institute in Sweden.
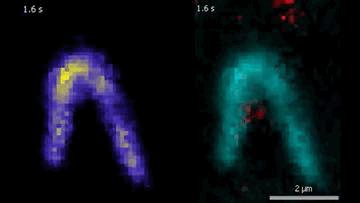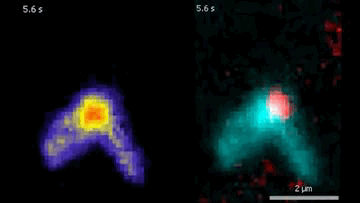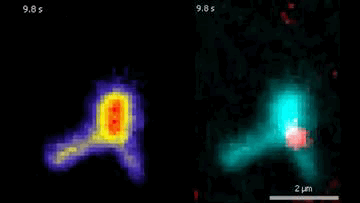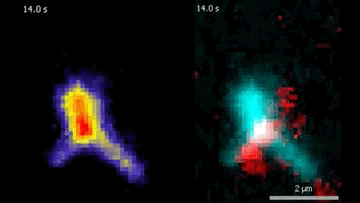
The DNA is labeled with fluorescent dyes and tethered to a glass surface on both ends to observe it in a microscope. The videos show how the Protein Smc5/6 extrudes DNA loops in real-time. In the left video, it can be seen that the Smc5/6 protein (red) binds to the DNA (cyan) and then starts to extrude the loop.
© Biswajit Pradhan / Max Planck Institute of Biophysics
Reproduction and evolution of life – from humans to bacteria – are based on preserving, adapting and passing on genetic information. The genetic material is stored in the form of DNA in almost every single cell of an organism, or more precisely in the nuclei. The cell nucleus is only a few millionths of a millimeter in size but contains DNA of about 2 meters length. Twisted space-savingly, the threadlike DNA forms the characteristic X-shaped chromosomes that we find in biology textbooks. During the cell’s life cycle, the chromosomes must be continuously loosened and compacted: During gene expression – i.e. the conversion of genetic information into physical characteristics or biochemical processes – or cell division, proteins have to dock to the DNA and migrate along it in order to read-out information or duplicate the DNA. If these processes do not function properly, rare hereditary diseases, developmental disorders, or cancer can emerge.
The Structural Maintenance of Chromosomes (SMC) protein complexes condensin, cohesin and Smc5/6 play a central role in the spatial organization of chromosomes. “We have already studied two major players in chromosome organization, condensin and cohesin. They extrude loops from DNA strands and hold them together like an eyelet through their ring-like structure, ” explains Eugene Kim, group leader at the Max Planck Institute of Biophysics. “Now, we have looked at the last puzzle piece Smc5/6 in DNA loop formation and unraveled its function. We found that the extrusion of DNA loops is a conserved mechanism in all eukaryotes, which are organisms with cell nuclei.
How does Smc5/6 extrude DNA loops?
By labeling DNA and Smc5/6 proteins with fluorescent dyes, the researchers were able to observe their interactions in real time under the microscope. They found that individual Smc5/6 proteins move unidirectionally along the DNA. Only when two Smc5/6 proteins assemble and form a dimer does the complex reel DNA into loops. You can think of this as pulling a loop out of a tangled rope and holding it together at the bottom with your hand so that two specific points of the rope touch. The proximity of specific DNA sections stimulates or suppresses processes such as gene expression or DNA replication.
A Smc5/6 protein consists of eight subunits. Two of them inhibit the formation of Smc5/6 dimers and, thus, regulate DNA-loop extrusion. In fact, if these two subunits are removed, looping of DNA continues almost endlessly, as the researchers observed. “We want to find out now how the process of loop formation is inhibited or stimulated in living cells. We will stepwise increase the complexity of our system in the microscope and see what happens,” says Biswajit Pradhan, one of the study’s first authors. “If we understand all details of DNA loop extrusion, it may be possible in the future to develop new therapies for diseases caused by defects in DNA organization.”
Successful collaboration
The project was a collaboration between Eugene Kim’s research group at the Max Planck Institute of Biophysics in Frankfurt am Main, Germany, and researchers at the Karolinska Institute in Sweden. Kim met Camilla Björkegren, group leader at the Karolinska Institute, during a scientific conference. “From the beginning, we were connected by the mystery surrounding the Smc5/6 complex,” Kim says.
Even though part of the experiments were conducted in Germany and the other in Sweden, the scientific exchange was exceptionally lively and ultimately brought success. “It was the best collaboration we ever had! Everyone was highly motivated and enthusiastic, and everyone contributed enormously to the intellectual progress of the project,” Kim points out.







