Researchers from Nano Life Science Institute (WPI-NanoLSI), Kanazawa University report in Nature Communications that TMEM16F, a transmembrane protein that facilitates the passive movement of phospholipids and ions across membranes, explores a larger conformational landscape than previously thought to perform its unique functions. The finding refines our molecular understanding of crucial physiological processes such as blood coagulation and COVID-19 pathogenesis, and highlights the importance of probing membrane proteins in native-like environments.
————————————————————————————————————————————————————————————————
Lipid scramblases are proteins embedded in cell membranes that play a crucial role in shuffling phospholipids between the two lipid layers that form such cellular boundaries. TMEM16F, a member of the TMEM16 protein family, acts as both a calcium-activated ion channel and a lipid scramblase, meaning that it can facilitate the transfer of both, lipids and ions across the chemical environment outside and inside of the cell. These movements regulate several biological functions such as blood clotting, bone development, and viral entry and are therefore of great physiological and clinical interest. At the molecular level, the TMEM16F architecture has a double-barrelled shape in which two identical polypeptide chains (called subunits), each formed by ten transmembrane (TM) helices, stick together (a process known as dimerization) to form two separate and presumably independent ion and lipid pathways.
Previously, it was thought that TMEM16F might work like a simple gate, with calcium ions serving as keys to unlock the two permeation pathways. Opening and closing the gate to different extents would let lipids and ions cross the plasma membrane alternately. However, structural investigations using cryo-electron microscopy (cryo-EM) -an in vitro technique that can reveal the 3D architecture of purified and frozen proteins at near-atomic resolution – have mostly captured TMEM16F snapshots in inactive conformations, with the ion and lipid gates presumably trapped in a closed state, raising questions about the validity of existing models.
To gain a better understanding of TMEM16F’s structure and function relationship, Holger Flechsig and Clemens Franz from WPI-NanoLSI, Kanazawa University, in collaboration with Vincent Torre from the International School of Advanced Studies (Italy) and former WPI-NanoLSI members Leonardo Puppulin and Arin Marchesi, used advanced techniques such as single-molecule force spectroscopy (SMFS) and high-speed atomic force microscopy (HS-AFM) imaging. These methods allowed them to observe TMEM16F behaviour at the molecular level in physiological environments, providing insights into its structure, dynamics, and mechanical properties.
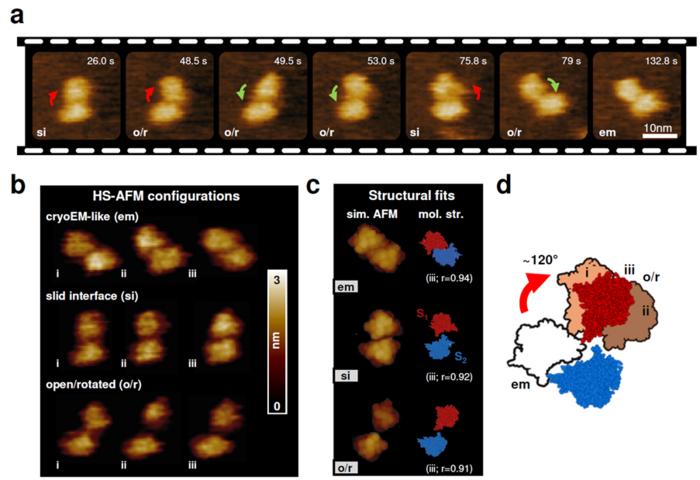
The study uncovered that TMEM16F exhibits a wide range of structural conformations that have been overlooked so far. The research revealed unexpected changes in the dimerization interface and TMEM16F subunit arrangements, suggesting that TMEM16F operates in a more dynamic and versatile manner than previously thought (Figure 1). The authors propose that these large structural variations are critical for TMEM16F’s diverse functions, including lipid scrambling and ion movement across the cell membrane. Furthermore, the researchers also found that calcium binding leads to significant rearrangements in specific regions of the protein, in particular in the transmembrane helices TM3, TM4, and TM6, which may lead to the opening of the ion and lipid pathways.
Overall, the research extends previous structural studies, demonstrates the complexity of the TMEM16F’s structure-function relationship, and highlights the importance of probing membrane proteins in native-like environments. Understanding these structural nuances could pave the way for targeted therapies and interventions to modulate TMEM16F activity in various diseases and physiological conditions.
Fig 1:
HS-AFM images of a murine TMEM16F dimer (a), main dimer configurations observed (b), and corresponding structural fits (c), highlighting the movement of the two subunits S1 and S2 (d).
© 2024 Zhongjie Ye, et al
Reference
Authors: Zhongjie Ye, Nicola Galvanetto, Leonardo Puppulin, Simone Pifferi, Holger Flechsig, Melanie Arndt, Cesar Adolfo Sánchez Triviño, Michael Di Palma, Shifeng Guo, Horst Vogel, Anna Menini, Clemens M. Franz, Vincent Torre & Arin Marchesi
Title: Structural heterogeneity of the ion and lipid channel TMEM16F
Journal: Nature Communications
Published on Jan. 2, 2024.
DOI: 10.1038/s41467-023-44377-7
URL: https://doi.org/10.1038/s41467-023-44377-7